This film of a 10 nanosecond simulation of molecular dynamics shows how the shape of a SARS-CoV-2 The virus protein (magenta) changes when it interacts with a possible low molecular weight inhibitor (blue).
Students conduct computer studies and research inhibitors to destroy viral proteins that help infectious particles escape from cells.
Detailed knowledge of how SARS-CoV-2, the virus that causes it COVID-19, replicating, and how the body reacts can suggest various strategies to stop it. Many researchers have worked to block the coronavirus spike protein from interacting with the receptors for human cells it binds to – the first step in infection. In contrast, three high school research program students who participated in research this summer with scientists from the Computational Science Initiative at the US Department of Energy’s Brookhaven National Laboratory sought one of the final steps – the virus’s exit strategy.
“After the virus is replicated and put together, it has to leave the cell,” said Peggy Yin, an aspiring senior at Port Jefferson High School. “Our body has an immune response in the form of a protein called ‘tetherin’ that binds the newly replicated virus particles to the cell membrane so that they cannot be free to infect other cells. This is a really useful tactic that our body has built in to keep us safe. ”
Unfortunately, SARS-CoV-2 has a way around this defense. The virus makes a protein that disrupts the tethering protein. “So if we inhibit the viral protein, we can let ‘tetherin’ do its thing,” said Yin.
The first step was to learn more about how the virus protein worked.
Peggy Yin of Port Jefferson High School at her home office. Photo credit: Brookhaven Lab
Modeling molecular interactions
Yin and his HSRP fellow students Jacob Zietek and Christopher Jannotta, who had just graduated from high schools in Farmingdale and Eastport South Manor, conducted protein-protein docking studies to model how the viral protein and the tethering protein interact .
“With this program, we can see where these proteins speak to each other, where they bind to each other and how the virus actually inhibits tetherin,” said Jannotta.
The modeling studies confirmed a suggestion the students had read about in the literature: the viral protein binds to parts of the tether that are glycosylated (added sugar groups), a necessary step for tetherin to work.
“We know that glycosylation takes place in the endoplasmic reticulum, an internal organelle of the host cell,” said Jannotta. “That means that if we want to develop some kind of inhibitor of the virus protein, we may have to get it into this internal organelle. But at least we now knew where to look for possible inhibitors on the viral protein. ”

Christopher Jannotta, a 2020 Eastport South Manor High School graduate, works from home. Photo credit: Brookhaven Lab
Look for inhibitors
The students conducted additional docking studies – this time looking at the interactions of the virus protein with many possible small drug-like molecules, or “ligands,” to find out which ones might be preventing the virus from blocking glycosylation.
“These protein-ligand docking studies are trying to put in the pocket the tiny ligands that are blocking glycosylation in order to find out which and which conformation or“ pose ”of the ligand best binds to the pocket,” Yin said. Based on 60 candidate ligands, the students narrowed the search to six or seven.
Then the team went further and performed molecular dynamic simulations of these ligand candidates. As Zietek explained, molecular dynamics simulations predict how the shapes of the protein and ligand will adapt and change over time, rather than just predicting whether a particular ligand will fit in a pocket of the protein – like a key that fits in a lock .
“These are much more complicated to calculate,” said Zietek, because it depends on what happens to individual atoms. “The program will calculate all the forces of atoms interacting with other atoms and will change the positions of atoms relative to one another over time to get as close as possible to what would happen in real life.”
Jacob Zietek, 2020 graduate of Farmingdale High School, in a robotics competition last year. Photo credit: Brookhaven Lab
Supercomputing power
“Because the project these students were working on contributes to a broader collaboration – the National Virtual Biotechnology Laboratory (NVBL) – they had access to Brookhaven’s supercomputer clusters to run the molecular dynamics simulations,” said Hubertus Van Dam, their mentor .
This is important because the molecular interactions between each ligand and the viral protein are only followed for 10 nanoseconds – 10 billionth of one second – takes 8-12 hours even on such powerful machines. “It would take way too long on normal computers!” Zietek noticed.
Ten nanoseconds doesn’t seem like a lot of time, but the simulations capture what happens every two seconds Femtoseconds– Millionths of a billionth of a second, noted Jannotta. “The computer is slowing it down so we can see it in real time,” he said.
“It’s like a slow motion camera,” added Yin.
As the team found out through the first, even shorter simulations, 10 nanoseconds are long enough to “find out whether a ligand is stuck in a protein or torn off,” said Zietek.
The students have quantified the results of the molecular dynamics simulations and are working on the best potential inhibitors and identifying which parts of the small drug-like molecules bind best to the viral protein. They are also looking for candidates to inhibit the viral protein in other ways. These initial computer studies pave the way for future experiments – and could even lead other scientists, most likely in pharmaceutical companies, to develop these ideas into actual drugs to inhibit SARS-CoV-2.
“I definitely think we’re going to help all the scientists working on these therapeutics,” said Jannotta, who will start biomedical engineering and initial studies at Stony Brook University this fall. “It will help them get a closer look at which inhibitors might actually be worth pursuing in pharmacology. Other researchers, perhaps in pharmaceutical companies, could pick it up and move on. I’m very excited for that. ”
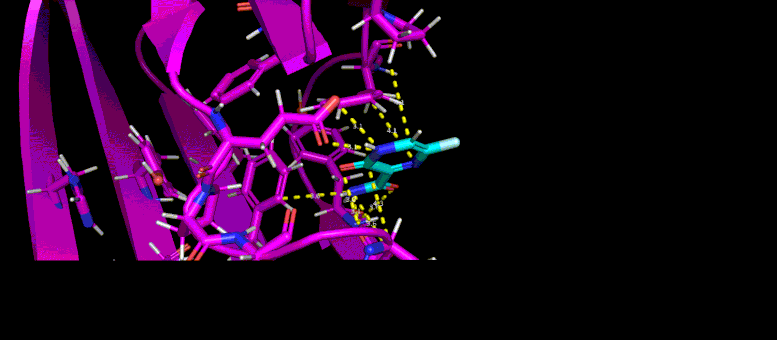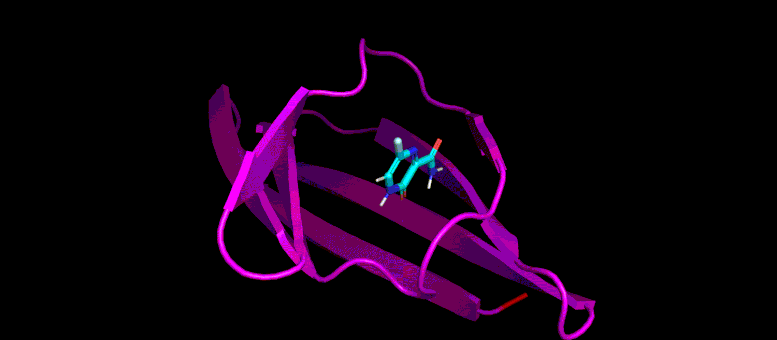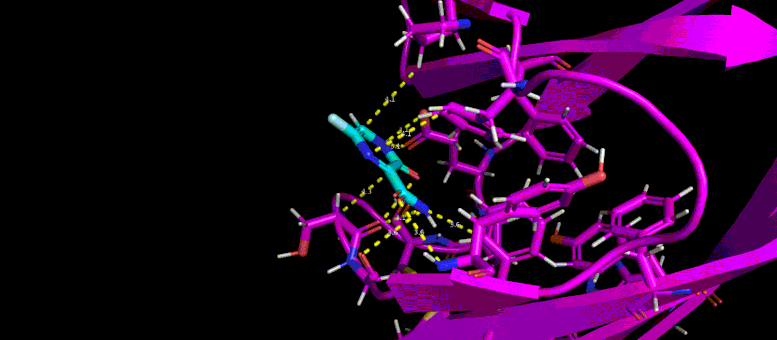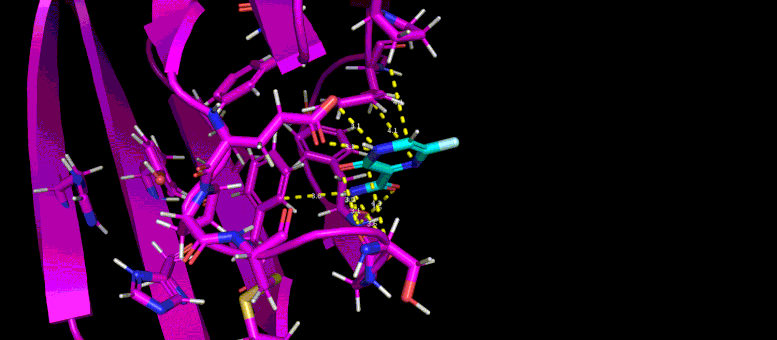
This GIF shows the predicted docking position of a possible low molecular weight inhibitor (blue) in a target pocket on a SARS-CoV-2 virus protein (magenta). Photo credit: Brookhaven Lab
Virtual is reality
Van Dam commented, “This is real research with real potential effects. The fact that these internships were “virtual” really made no difference. During the pandemic, even our professional computer scientists conducted similar computer studies carried out from home. ”
Zietek, who will be attending Purdue University in the fall, said: “I never thought I would be working on such a relevant and urgent topic as COVID-19 when I first applied for the Brookhaven program. I knew I wanted to work on a computational project to learn more about how computers could be used in a research setting. But this was almost the most emotional subject you could get for a project. I was very excited to have the opportunity to contribute. ”
Yin was in the COVID angle from the start.
“When I applied in January, I mentioned in my essay that I wanted to do computational biology research. And when the pandemic situation worsened, I emailed the possibility that I could do COVID-19 research because I really wanted to help, ”she said. “I hope that, since we know so little about coronaviruses in general, our research may shed some light on how these viruses work, potentially help in other areas of coronavirus research, and prevent other pandemics in the future. ”
Modeling of CSI virus protein / drug development is supported by the DOE Office of Science (BER) through the National Virtual Biotechnology Laboratory (NVBL), a consortium of national DOE laboratories focused on responding to COVID-19 and supported by the EU is funded by the Coronavirus CARES Act. Student participation in this project was supported by HSRP, a program run by the Brookhaven Lab Educational Programs Office with funding from Brookhaven Science Associates – a partnership between Battelle and the State University of New York Research Foundation on behalf of Stony Brook University -. manages the Brookhaven Lab.



