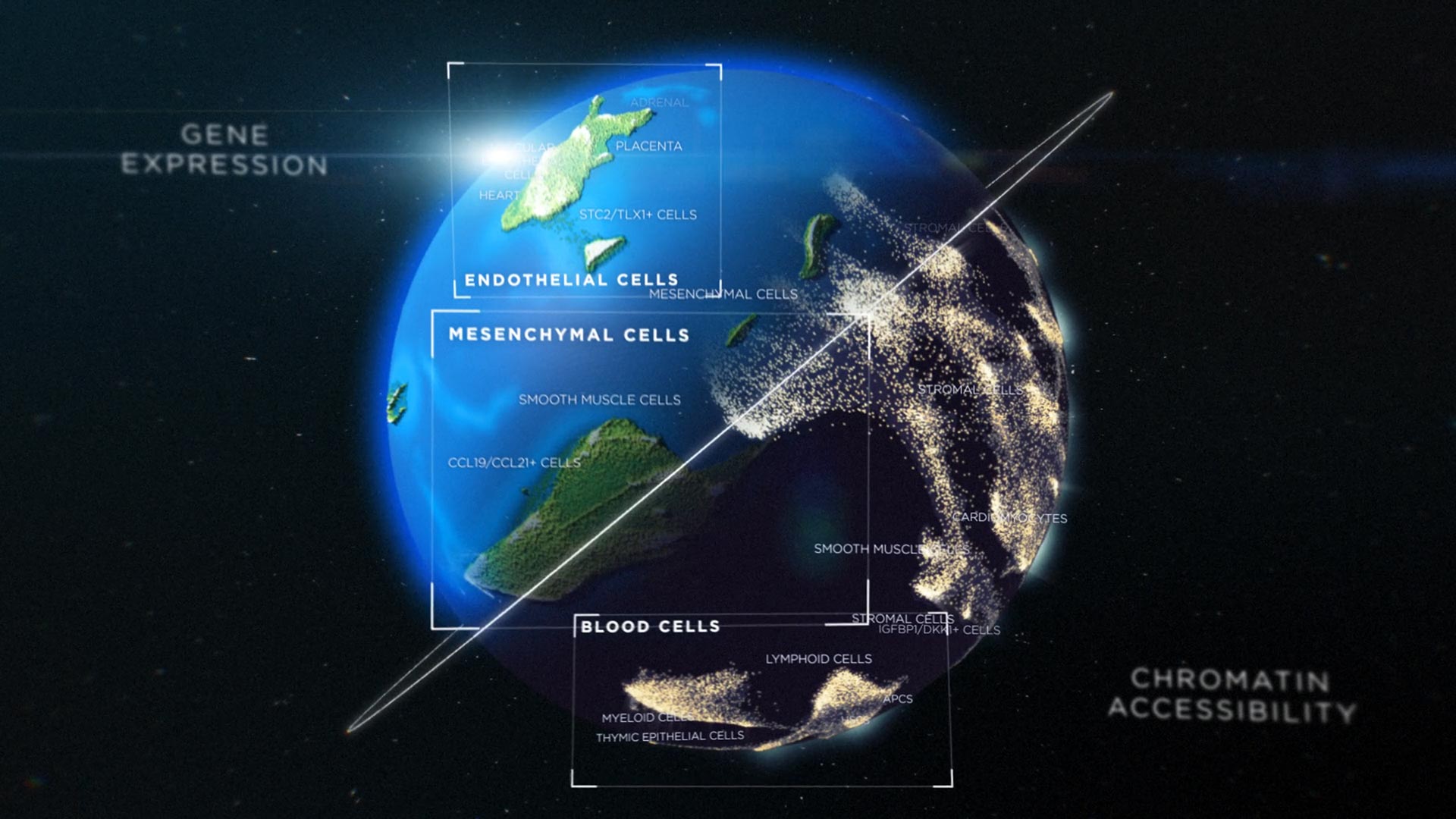
An artistic interpretation of the creation of two cell atlases that track gene expression and accessibility to chromatin during the development of human cell types and tissues. The atlases are reported in the November 12th issue of Science. They were developed by the Brotman Baty Institute for Precision Medicine and Underwater Medicine in Seattle. Photo credit: Dani Bergy and Inessa Stanishevskaya / Cognition Studio, Inc.
The two atlases depict gene expression and availability when cells differentiate into different cell types and tissues.
Researchers from UW Medicine at the Brotman Baty Institute in Seattle have developed two cell atlases that track gene expression and accessibility to chromatin during the development of human cell types and tissues.
An atlas depicts gene expression within individual cells in 15 fetal tissues. The second atlas depicts the chromatin accessibility of individual genes within the cells.
Together, the atlases provide a fundamental resource for understanding gene expression and accessibility to chromatin in human development that is unprecedented in scale. In addition, the techniques described in the two publications enable the generation of data on gene expression and the availability of chromatin for millions of cells.
The atlases are described in successive publications in the November 13th issue of the journal science. In addition to BBI and UW Medicine, other employees from Illumina, Inc., the University of Arizona, the Fred Hutchinson Cancer Research Center, the Max Planck Institute for Molecular Genetics, and the University of Rochester Medical Center contributed to these studies.
Gene Expression Atlas
Gene expression is the process by which a cell uses the instructions stored in its cell DNA Control protein synthesis. These proteins in turn determine the structure and function of a cell. The gene expression atlas maps where and when gene expression occurs in different cell types as they grow and develop.
“From this data we can directly compile a catalog of all the major cell types in human tissues, including how these cell types can vary in gene expression in different tissues,” said lead author Junyue Cao. He completed this work as a postdoctoral fellow in the laboratory of Jay Shendure, professor of genomic science at UW Medicine, researcher at Howard Hughes Medical Institute and scientific director of the Brotman Baty Institute. Cao is now an assistant professor at Rockefeller University.
“It is an overarching goal of the field to profile the genetic programs that exist in a human on the broadest possible scale and with as much resolution as possible,” Shendure said.
To create the atlas, the researchers profiled gene expression across 15 types of fetal tissue using a technique called sciRNA-seq3. This technique marks each cell with a unique combination of three DNA “barcodes”, which allows researchers to track the cells without physically separating them.
Once the sequences were obtained, they used computer algorithms to recover the single cell information, group cells by type and subtype, and identify their evolutionary trajectories. The scientists profiled over 4 million individual cells and identified 77 major cell types and approximately 650 cell subtypes.
They also compared the atlas to an existing atlas of mouse embryonic development. Co-senior author Cole Trapnell, associate professor of genomic science at the UW School of Medicine and researcher at the Brotman Baty Institute, stated, “When we combine this data with previously published data, we can directly describe the cell’s evolutionary path for all major cells Types. ”
DNA Accessibility Atlas
The second atlas of DNA accessibility maps material called chromatin, which can be used to pack DNA tightly into the nucleus. Chromatin can be open and “accessible” to the molecular machinery that reads the genetic instructions encoded in DNA, or closed and “inaccessible”. Knowing open and closed regions of DNA can help you see how a cell turns genes on and off.
Studying chromatin gives you an idea of the cell’s regulatory “grammar,” said co-senior author Darren Cusanovich, previously a postdoctoral fellow in the Shendure laboratory and now an assistant professor at the University of Arizona. “The short stretches of DNA that are open or accessible are enriched for specific ‘words’, which in turn are the basis for specifying the cell on which particular genes are to be identified.”
In order to examine the accessibility of DNA in individual cells, the scientists developed a new method called sci-ATAC-seq3. As with sci-RNA-seq3, this technique also uses three different DNA barcodes in each cell to mark and track individual cells. However, instead of identifying all currently expressed sequences, sci-ATAC-seq3 captures chromatin sites and sequencing them.
In this study, the scientists created nearly 800,000 single cell chromatin accessibility profiles in approximately 1 million locations in 15 fetal tissues. They asked which proteins are likely to interact with accessible DNA sites in each cell and how these interactions may explain the cell type. This analysis defines the control switches for development within the genome. They also identified chromatin accessibility sites that could be linked to disease.
“This tells us which part of the genome might be functional. We still don’t know what percentage of the genome that doesn’t code for genes can be involved in gene regulation. Our atlas now provides this information for many cell types, ”said Silvia Domcke, co-lead author of the paper on accessibility and a postdoc in the Shendure laboratory. The other lead authors on the DNA accessibility study are Andrew Hill, previously a computational biologist in Shendure’s lab and now a scientist at 10x Genomics, and Riza Daza, a scientist at Shendure’s lab.
References:
“A Human Cell Atlas of Fetal Gene Expression” November 12, 2020, science.
DOI: 10.1126 / science.aba7721
“A Human Cell Atlas of Chromatin Accessibility” November 12, 2020, science.