The artistic interpretation of how microbial genome sequences from the GEM catalog can help fill knowledge gaps about the microbes that play a key role in Earth’s microbiomes. This image complements a public repository of 52,515 microbial design genomes generated from environmental samples around the world, adding 44% to the known diversity of bacteria and archaea. It is now available and will be described in Nature Biotechnology on November 9, 2020. The work on this catalog of genomes from the Earth’s microbiome catalog is the result of a collaboration between more than 200 scientists, researchers from the DOE Joint Genome Institute (JGI) and the DOE Systems Biology Knowledgebase (KBase). Photo credit: Rendered by Zosia Rostomian / Berkeley Lab
Despite the advances in sequencing technologies and computational methods over the past decade, researchers have discovered genomes for only a small fraction of the Earth’s microbial diversity. Since most microbes cannot be cultivated in laboratory conditions, their genomes cannot be sequenced using conventional approaches. Identifying and characterizing the planet’s microbial diversity is key to understanding the role microorganisms play in regulating nutrient cycles, as well as gaining insight into potential uses they may have in a variety of research areas.
A public repository of 52,515 design microbial genomes generated from environmental samples around the world, expanding the known diversity of bacteria and archaea by 44%, is now available and will be described in November 9, 2020 Natural biotechnology. Known as the GEM (Genomes from Earth’s Microbiomes) catalog, this work is the result of a collaboration between more than 200 scientists, researchers from the Joint Genome Institute (JGI) of the US Department of Energy (DOE), a user facility of the DOE Office of Science at Lawrence Berkeley National Laboratory (Berkeley Lab) and the DOE Systems Biology Knowledgebase (KBase).
Metagenomics is the study of the microbial communities in the environmental samples without the need to isolate individual organisms using various methods for processing, sequencing and analysis. “Using a technique called metagenome binning, we were able to reconstruct thousands of metagenome-composite genomes (MAGs) directly from sequenced environmental samples without having to cultivate the microbes in the laboratory,” noted Stephen Nayfach, the study’s first author and scientist in The Microbiome Data Science group of Nikos Kyrpides. “What really sets this study apart from previous efforts is the remarkable environmental diversity of the samples we analyzed.”
Emiley Eloe-Fadrosh, director of the JGI metagenome program and lead author on the study, provided Nayfach’s comments. “This study is designed to cover the broadest and most diverse range of samples and environments, including natural and agricultural soils, human and animal-associated hosts, and marine and other aquatic environments – which is quite remarkable.”
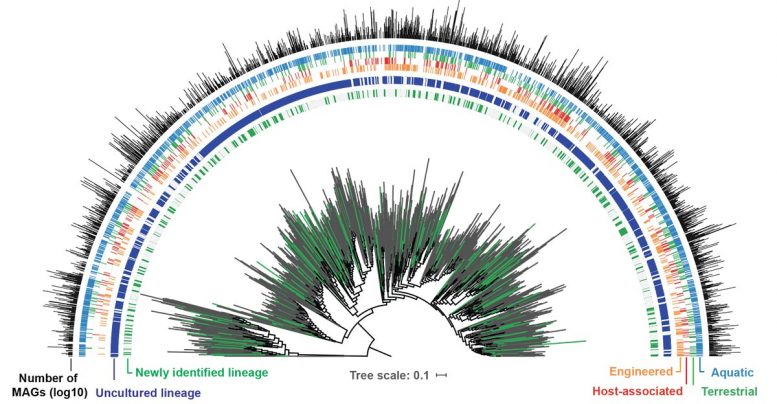
The GEM catalog extends the bacterial and archaic orders of the phylogenetic tree with new lineages of uncultivated genomes from the GEM catalog (in green) and previously existing reference genomes (in gray). Around the phylogenetic tree, the strip diagrams indicate whether an order is uncultivated (blue; represented only by metagenome-composite genomes or MAGs) or cultivated (gray; represented by an isolate genome). The next four strip charts show the environmental distribution, while the bar chart shows the number of genomes from the GEM catalog obtained from each order. Photo credit: Stephen Nayfach
Added value beyond genome sequences
Much of the data was generated from environmental samples that were sequenced by the JGI as part of the Community Science Program and was already available on the JGI’s IMG / M (Integrated Microbial Genomes & Microbiomes) platform. Eloe-Fadrosh noted that it was a fine example of “big data” mining to gain a deeper understanding of the data and add value by making data publicly available.
In recognition of the efforts of the investigators who carried out the sampling, Eloe-Fadrosh reached out to more than 200 researchers around the world, under the JGI Data Usage Policy. “I felt it was important to acknowledge the significant efforts made to collect and extract DNA From these samples, many of which come from unique, inaccessible environments, these researchers were invited to be co-authors in the IMG data consortium, ”she said.
Using this massive data set, Nayfach grouped the MAGs into 18,000 candidate species groups, 70% of which were new, compared to over 500,000 genomes available at the time. “When you look over the tree of life, you see how many uncultivated lineages are only represented by MAGs,” he said. “Even though these design genomes are imperfect, they can reveal a lot about the biology and diversity of uncultivated microbes.”
Research teams worked on several analyzes using the genome repository, and the IMG / M team developed several updates and features to break down the GEM catalog. (For more information, see this IMG webinar on Metagenome Bins.) One group examined the novel secondary metabolites data set from Biosynthetic Gene Clusters (BGCs) of secondary metabolites and increased these BGCs in IMG / ABC (Atlas of Biosynthetic Gene Clusters) by 31%. (Listen to this JGI Natural Prodcast episode on Genome Mining.) Nayfach also worked with another team on predicting host virus links between all viruses in IMG / VR (Virus) and the GEM catalog, wherein 81,000 viruses have been assigned – 70% of which has not already been connected to a host – with 23,000 MAGs.
Modeling a New Path for Metagenomists
Building on these resources, KBase, a multi-institutional environment for collaborative knowledge creation and discovery developed for biologists and bioinformaticians, developed metabolic models for thousands of MAGs. The models are now available in a public narrative that provides shareable, reproducible workflows. “Metabolic modeling is a routine analysis for isolate genomes, but has not been done on a large scale for uncultured microbes,” said Eloe-Fadrosh. “We believed that working with KBase would add value beyond clustering and analyzing these MAGs.”

The study used IMG data from environmental samples collected in the dry valleys of Artarctica. Photo credit: Craig Cary, International Center for Antarctic Terrestrial Research, University of Waikato
“Bringing just this dataset into KBase is of immediate value as people can find the high quality MAGs and use them for future analysis,” said José P. Faria, a KBase computational biologist at Argonne National Laboratory. “The process of building a metabolic model is simple: you simply select a genome or MAG and press a button to build a model from our database with associations between biochemical reactions and annotations. We look at what has been commented on in the genome and the resulting model to assess the organism’s metabolic capabilities. “(Check out this metabolic modeling KBase webinar.)
Elisha Wood-Charlson, director of KBase User Engagement, added that metagenomists may consider branching out into this area by demonstrating the ease with which metabolic models are generated from the GEM dataset. “Most metagenomics researchers may not be ready to dive into a whole new field of research [metabolic modeling], but they might be interested in how biochemistry affects what they are working on. The genomics community can now study metabolism using the simple KBase pathway of genomes or MAGs for modeling that may not have been considered, ”she said.
A community resource to facilitate research
Kostas Konstantinidis of the Georgia Institute of Technology, one of the co-authors whose data was part of the catalog: “I don’t think there are many institutions that can do this type of metagenomics on a large scale and have the capacity for large-scale metagenomics analyzes. The beauty of this study is that it is done on a scale that individual laboratories cannot, and it gives us new insights into microbial diversity and function. ”
Data in IMG from algae samples were used for the study. Photo credit: Erica Young
He is already finding ways to use the catalog in his own research on how microbes respond to climate change. “With this data set, I can see where each microbe is and how often it is present. This is very useful for my work and for others doing similar research. “In addition, he is interested in expanding the variety of the reference database he has developed called the Microbial Genomes Atlas to allow for more robust analyzes by adding the MAGs.
“This is a great resource for the community,” added Konstantinidis. “It is a data set that will enable many more studies later. And I hope that JGI and other institutions will continue to do these types of projects. ”
Reference: “A Genomic Catalog of the Earth’s Microbiome” by Stephen Nayfach, Simon Roux, Rekha Seshadri, Daniel Udwary, Neha Varghese, Frederik Schulz, Dongying Wu, David Paez-Espino, I-Min Chen, Marcel Huntemann, Krishna Palaniappan and Joshua Ladau Supratim Mukherjee, TBK Reddy, Torben Nielsen, Edward Kirton, José P. Faria, Janaka N. Edirisinghe, Christopher S. Henry, Sean P. Jungbluth, Dylan Chivian, Paramvir Dehal, Elisha M. Wood-Charlson, Adam P. Arkin Susannah G. Tringe, Axel Visel, IMG / M-Datenkonsortium, Tanja Woyke, Nigel J. Mouncey, Natalia N. Ivanova, Nikos C. Kyrpides and Emiley A. Eloe-Fadrosh, November 9, 2020, Natural biotechnology.
DOI: 10.1038 / s41587-020-0718-6
The work also used resources from the National Energy Research Scientific Computing Center (NERSC), another user facility of the DOE Office of Science at the Berkeley Lab.